CroPyDB
Crop phenomics requires the handling of large datasets which are being decomposed from massive raw data (such as hyperspectral images or point clouds of terrestrial laser scanners) to derive a final crop model predicting the targeted trait (mostly yield and its stability). Between these two ends, a range of different modelling steps is necessary. Typically, there is a need to train feature extraction models using ground truth, correct for design factors and spatial trends in the field, derive parameters from repeated measures (e.g. growth dynamics) and model the response to environmental covariates. Finally, there is the need to combine multiple traits derived from different sensors at different year-sites into a genetic crop model to support the selection of genotypes or markers. Without proper data handling this workflow in impossible to accomplish.
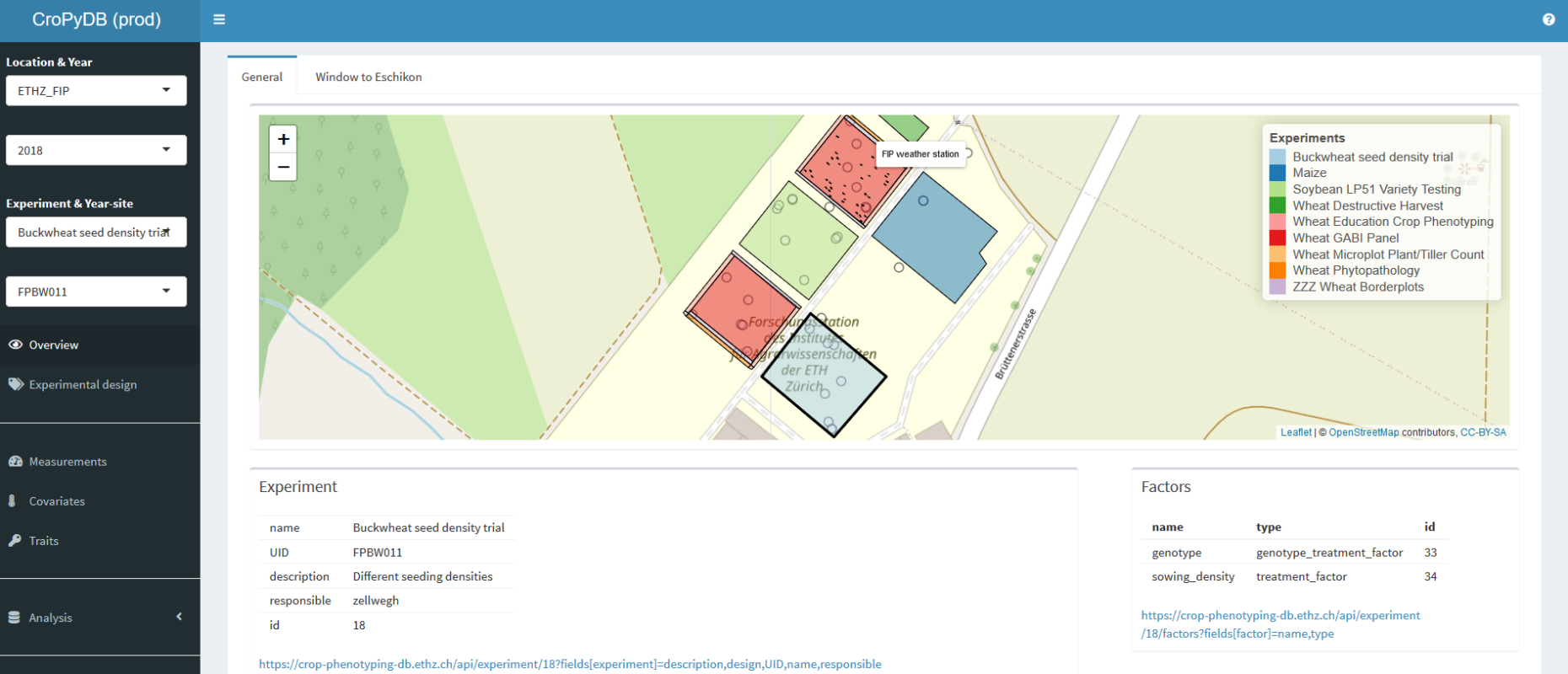
CroPyDB is being developed by the group of Crop Science to support data management in crop phenotyping and physiological breeding. It is written in external pagePython 3.6call_made and based on a `external pagePostgreSQL databasecall_made with the spatial data extension external pagePostGIScall_made. Crucial modules include external pageSQLAlchemycall_made as external pageObject-relational mappercall_made and external pageFlask-Restlesscall_made as external pageJSON-APIcall_made external pageRESTcall_made Server.
CroPyDB has two main aims:
- Support the collection and processing of phenomics data in our research group
- Ensure compatibility of our research data to standards in plant phenomics
Therefore, we ensured that the design of CroPyDB respects the requirements of external pageMIAPPEcall_made, links to the external pagecrop ontologycall_made and uses the external pageFAO standard on multi passport datacall_made.
Contact
Professur für Kulturpflanzenwiss.
Universitätstrasse 2
8092
Zürich
Switzerland
